Data and Model Version Control in ML Drug Discovery
05 December 2022
WWhen I first started applying ML to drug discovery, I quickly realized that messy biological data could break even the most elegant models. Tracking datasets, preprocessing steps, and models became just as important as building the algorithms themselves.
In my talk titled Applications in ML Drug Discovery Pipelines at PyData NYC 2022, I walked through how at Cyclica, I addressed ML experiments and reproducibility in Drug Discovery pipelines with DVC. In this blog post, I’ll share key themes from that journey, the lessons learned at Cyclica, and how those lessons might translate beyond drug discovery into any complex ML workflow.
The core problem: discovery is messy
Drug discovery is not like classifying images or predicting clicks. In our world, the data is not clean or standardized from the source, feedback is delayed, the scale is enormous, and the stakes are real. I found datasets that spanned millions of molecules, dozens of targets, highly variable assay conditions, and pipelines that had grown organically rather than by design.
Our team faced three intertwined issues early on: variation in how assays were run; scale in how many molecules and features needed to be processed; and the maturity of the pipelines themselves. If any of these break down, discovery slows, costs escalate, and risk rises.
A key step in working with data for drug discovery is cleaning, sanitizing, and standardizing it. Reproducibility becomes critical here: having a clear record of how data was processed, transformed, and used in model training is essential for trust and validation. This is where Data Version Control (DVC) comes into play.
Biological data is complex
In drug discovery, the basis for ML pipelines is biological and chemical data. Biological data is inherently complex because it reflects studies of intricate phenomena where many variables interact. Expecting simple, unidimensional data is unrealistic. Instead, biological data is heterogeneous, conditional, high-dimensional, and research-biased.
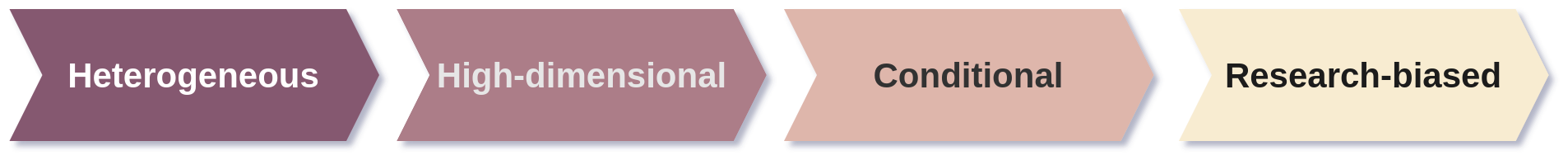
Here is why:
- Heterogeneous: Molecular biology has developed specialized data-driven methods and technologies to characterize and quantify biological systems at different levels: genomics, proteomics, transcriptomics, and metabolomics among others. Each derived from specific experimental essays and are reported in different formats.
- Conditional: To develop ML algorithms in drug discovery ground truths are a must and they are determined by reported experimental measurements. However, reproducing experiments may result in different, and sometimes conflicting outcomes. For example, running the same protocol in two different labs may lead to different results, and ground truths may be elusive.
- High-dimensional: In biological data, the number of samples (observations) is often limited and much fewer than the number of variables (features) due to limited available resources. For example, human genome expression arrays can probe for the expression of c.a. 47,000 transcripts in a single sample, meaning that the number of variables are five orders of magnitude higher than the number of samples!
- Research-biased: Chemical and biological findings are usually focused or biased towards specific outputs. It is more likely to find more data in areas with higher market value or on topics that are considered ‘hot’. A remarkable example of this bias can be found in the increased scientific production of COVID-19-related studies between December 2019 and April 2020.
In short, applying a “black box” model is not enough; the entire pipeline, from data versioning to experiment tracking to deployment, must be built with these constraints in mind.
Using Version Control for Data and Models
In software engineering, version control systems like Git track changes in code, enabling collaboration and reproducibility. In ML, particularly in data-intensive fields like drug discovery, versioning datasets and models is equally critical.
Data Version Control (DVC) brings Git-like tracking to datasets, preprocessing steps, and model artifacts, ensuring that experiments can be reproduced, audited, and shared across teams.
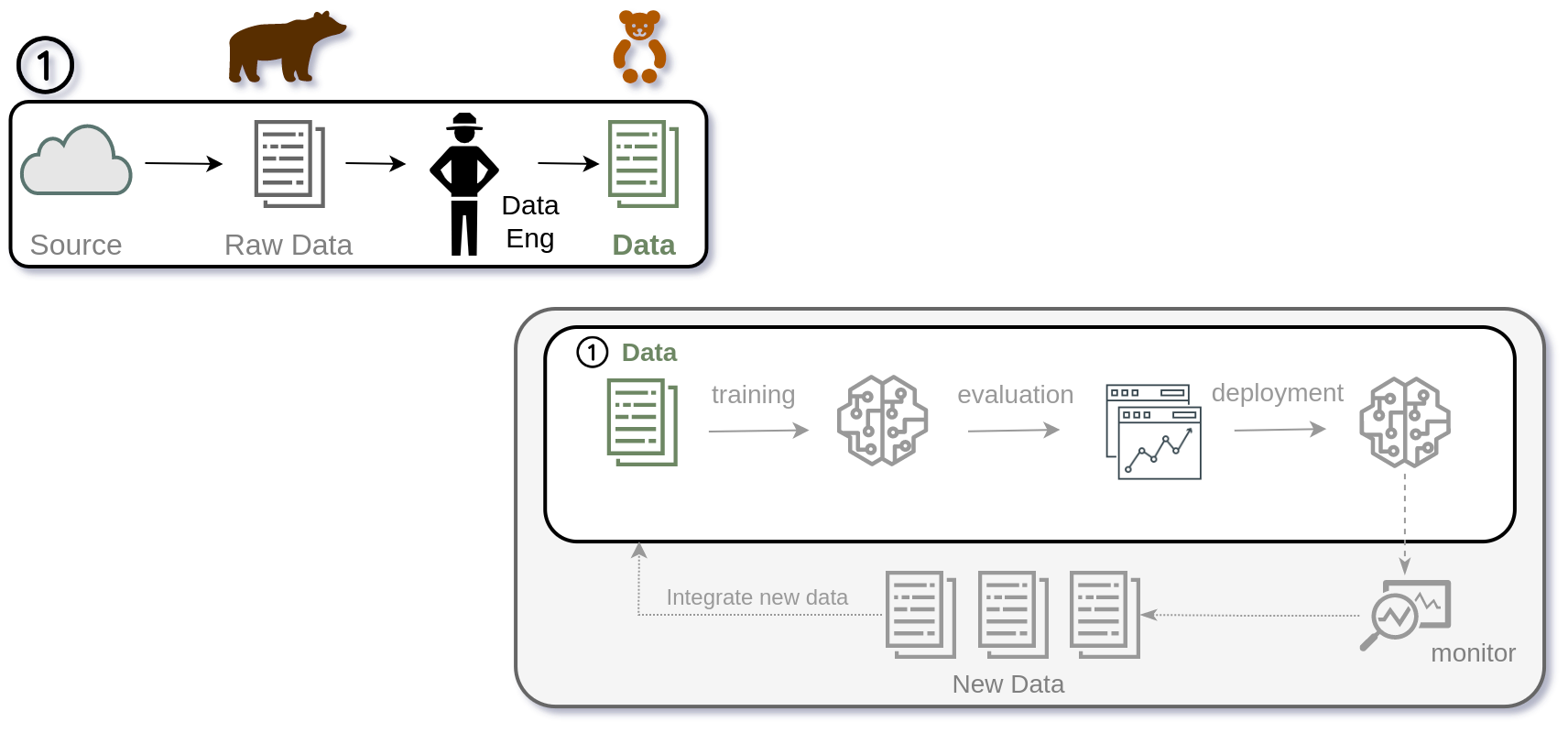
The first step in our journey was to implement DVC to version control datasets, preprocessing scripts, and model artifacts. This allowed us to track changes over time, revert to previous versions, and ensure that experiments were reproducible.
The second step was to integrate DVC for experiment tracking. This enabled us to log parameters, metrics, and outputs for each experiment, making it easier to compare different model versions and configurations.
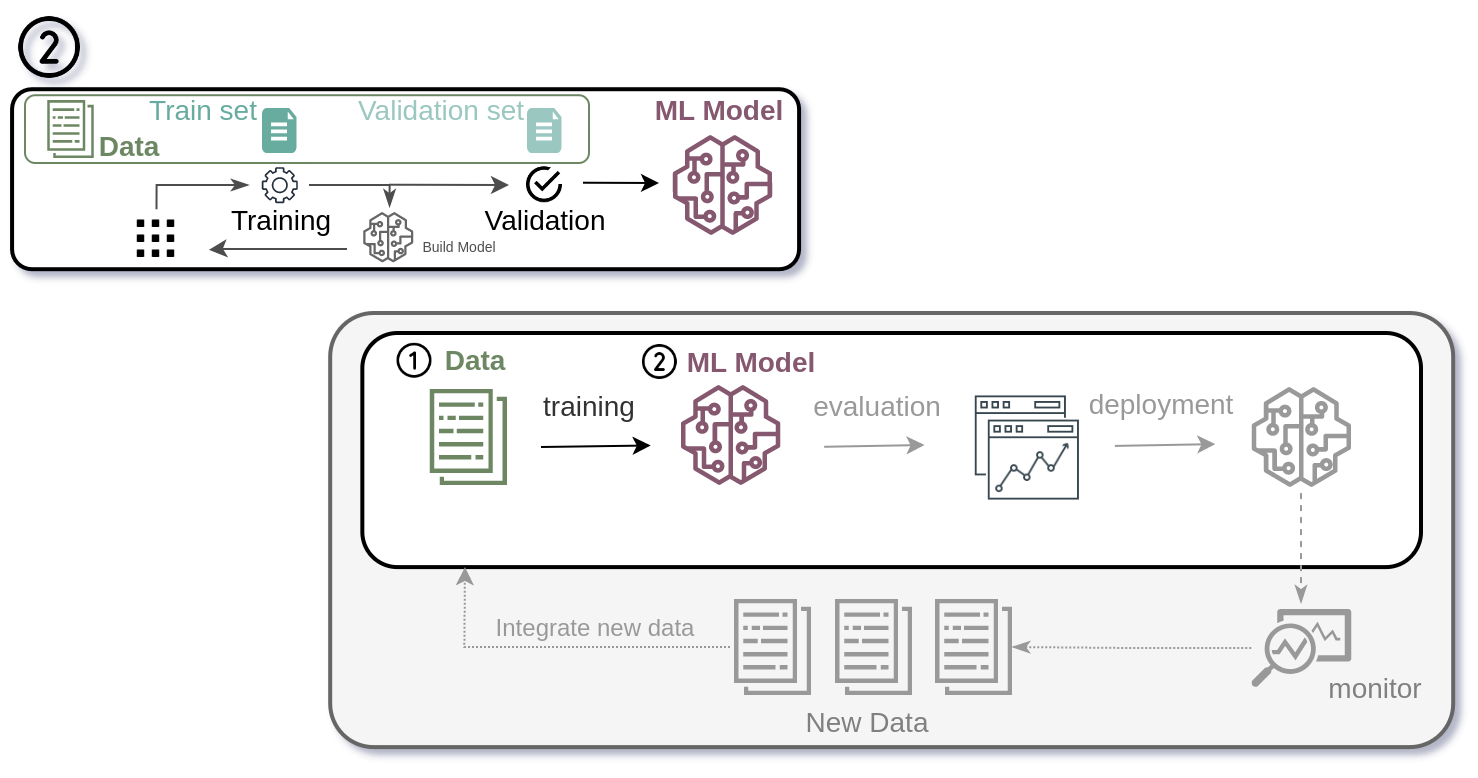
Once the cleaned data reaches the training stage, the real challenge begins. Model training in drug discovery is computationally heavy, often relying on deep learning architectures that require multiple rounds of feedback, tuning, and virtual screening. After many cycles of training and optimization, we can produce a model suitable for deployment, often as an API or within a larger application. Yet even then, success is fleeting: new assays, compounds, and experimental results quickly render models obsolete. Maintaining accuracy demands continuous data updates, retraining, and vigilance, which makes reproducibility and version control essential not only for the model itself but for the entire pipeline.
Building the pipeline: from sketch to production
In many start-ups, ML development starts in notebooks. Data is explored, models are tested, and metrics are evaluated. Then, during a grow stage the question “Will this run tomorrow, will it scale, will someone reproduce it next week?” comes up.
This shift, from proof of concept to production pipeline, is where many projects stall or deliver limited value.
At Cyclica, I addressed these challenges by building a robust ML pipeline using Data Version Control (DVC) to manage datasets and models which also brought us the advantage of having experiment tracking, reproducibility, and collaboration across teams.
Here are the key components we considered:
- Data Versioning: I used DVC to version control datasets, preprocessing steps, and feature engineering processes. This allowed us to track changes over time, revert to previous versions, and ensure that experiments were reproducible.
- Experiment Tracking: DVC was used for experiment tracking tools to log parameters, metrics, and outputs. This enabled us to compare different model versions and configurations easily.
- Reproducible Environments: By combining DVC with Docker, we created isolated environments that preserved dependencies and configurations. This ensured that models could be reproduced across different machines and setups.
- Automated Workflows: I set up automated pipelines using DVC stages to streamline data processing, model training, and evaluation.

If you’re working on ML in a domain like drug discovery (or anything with high cost of error), here is a practical roadmap:
- Map your pipeline: Sketch the path from raw data to prediction to decision. Where are the gaps?
- Version your data: Start tracking dataset versions, preprocessing steps, who changed what.
- Track your experiments: Use a simple experiment tracking tool (even a spreadsheet is fine to start) and log parameters + outcomes.
- Define your true metric: Ask stakeholders: what matters most? What cost do false positives/negatives bring?
- Plan for maintenance: Set calendar reminders for pipeline review, drift checks, data refresh.
Even these five steps will place you already be ahead of many teams who built a model and then watched it quietly drift into obsolescence.
The payoff: models that don’t just live in notebooks but deliver value in biological labs, research teams and production systems.
Whether in drug discovery or any high-stakes ML domain, investing in reproducibility, versioning, and domain-aligned pipelines is the difference between fragile models and reliable, actionable insights.
These lessons extend beyond drug discovery: in any high-stakes ML domain, reproducibility, versioning, and pipeline robustness are non-negotiable for delivering real-world impact.
▶️ Watch the talk
The full talk from PyData NYC 2022 here. After this talk was posted by PyData NYC, I was contacted by Iterative team (the company that created DVC) to share the talk on LinkedIn, and make it available on the Iterative Youtube channel.
The slides are also available on GitHub.