The project
22 June 2021
A brief introduction to my GSoC project!
It’s been two weeks since I started my Google Summer of Code (GSoC) journey and before moving forward, I would like to take a moment to share details of my_project, the motivations behind it, as well as my expectations. The main goal is to integrate a membrane curvature analysis tool into MDAnalysis. This tool will enable users to calculate the membrane curvature, in terms of mean and Gaussian curvature, of biological membranes.
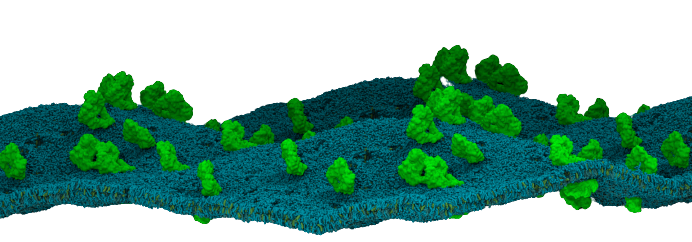
Fundamentally, biological membranes are functional and structural assemblies composed of a lipid bilayer matrix and associated proteins. Despite this simplification, biological membranes are diverse and highly complex. Their lipid composition varies according to their physical properties and functional roles. Additionally, their asymmetrical lipid composition by leaflet also contributes to maintaining specific mechanisms. Hence, biological membranes are not a pure architecture trick played by the cell. Instead, they actively drive many processes involving membrane remodeling and membrane trafficking.
- How does
membrane_curvaturework? (A glimpse)
To calculate membrane curvature, we first need a surface. Given the nature of biological membranes, we can use phospholipids -core elements of lipid bilayers-, as elements of reference to derive the surfaces associated with each leaflet. For simplicity, we can use the lipid head groups to define the surface. Once we think of leaflets as surfaces defined by lipid headgroups, we can calculate the mean and Gaussian curvature.
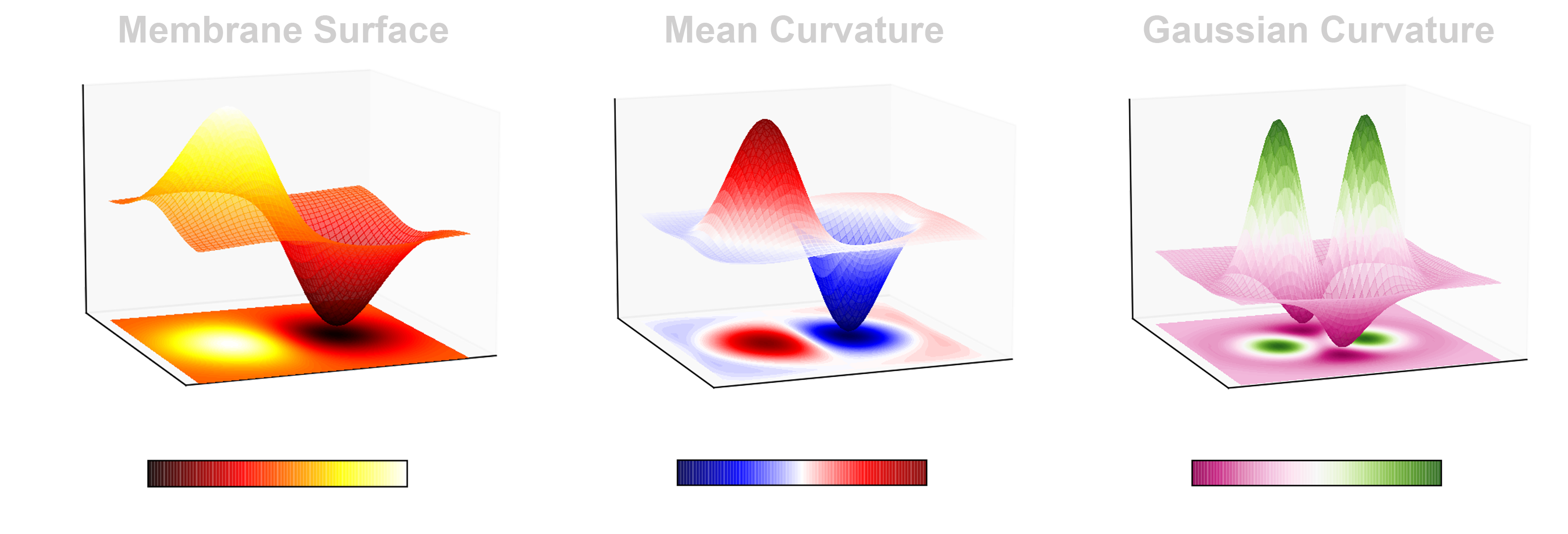
Mean curvature is an extrinsic property of a surface. Gaussian curvature, on the other hand, is intrinsic. That is to say, mean curvature depends on the coordinate space in which the surface is embedded, while the Gaussian curvature can be calculated within the surface itself without any reference to a larger space. Mean and Gaussian curvature together give information about how curved and how elastic the membrane is. In the figure below, given an arbitrary surface (left panel), the corresponding plots of mean (middle) and Gaussian curvature (right panel) are shown.
- What will this project unlock? What will the membrane curvature tool enable?
If you are in the MD simulations field and you work with any protein-membrane or membrane-only system, this tool will help your research work! Findings reported in literature suggest that there is an interplay between membrane curvature and protein sorting [1] [2] [3] and/or protein function [4][5]. Although this complex interaction may be determined by the specific features of the membrane (lipid type, lipid composition) or the protein (protein shape, type of residues facing the membrane, protein conformation), curvature of biological membranes can provide valuable insight when addressing a research problem.
- When and where will I find the
membrane_curvaturetool?
The membrane curvature tool is expected to be part of the MDAnalysis analysis package in future versions. A sample of how it is expected to work in the future looks like this
import MDAnalysis as mda
from MDAnalysis.analysis.curvature import Curvature
atom_file = <your coordinates file from MD simulations>
traj_file = <your trajectory from MD simulations>
u = mda.Universe(atom_file, traj_file)
surface = u.select_atoms("<atoms of reference>")
curvature = Curvature(surface).run()
mean = curvature.mean_curvature()
gaussian = curvature.gaussian_curvature()- What are the limitations to calculate membrane curvature?
When calculating membrane curvature in MD simulations, there are two main limitations.
The first one is sampling. To properly sample a decent range of curvatures in MD, the required simulation times are very large, reaching dozens or hundreds of microseconds. Such time scales are expensive and sometimes only affordable by state-of-the-art research groups.
The second limitation is the lack of user-friendly, actively-maintained and well-documented tools to analyze membrane curvature. Although some GitHub repos exist, not all of them are maintained and/or documented, some lack source code, benchmarks are non-existent, or they have not been ported to the modern Python 3 environment.
This was, in fact, one of my strongest motivations to contribute a membrane curvature tool under the MDAnalysis umbrella. With membrane_curvature, students and researchers will be able to calculate mean and gaussian curvature of biological membranes using one of the most popular, easy-to-use, and actively-maintained packages for analysis of MD simulations.
I started developing this tool as part of my Ph.D. and I would like to share it with the wider MD community. I am convinced that by sharing our knowledge and open-sourcing our algorithms and tools, science will move forward at a faster pace.
Share the code and help the world!

And while you are here, did you know that MDAnalysis is one of the most popular Python libraries to analyze MD simulations? MDAnalysis core developers actively work to make it better with every release. Please consider donating today.
References:
- [1] ACS Cent. Sci. 2020, 6, 7, 1159–1168.
- [2] Nat Commun 6, 8728 (2015).
- [3] Soft Matter, 2021,17, 4254-4265.
- [4] J Cell Sci (2015) 128 (6): 1065–1070.
- [5] eLife 2019;8:e50576.